How do we compare the scale of language learning input for large language models vs. humans? I've been trying to come to grips with recent progress in AI. Let me explain two illustrations I made to help.
Recent progress in AI is truly astonishing, though somewhat hard to interpret. I don't want to reiterate recent discussion, but
@spiantado has a good take in the first part of
lingbuzz.net/lingbuzz/007180; l like this thoughtful piece by
@MelMitchell1 as well:
https://www.pnas.org/doi/10.1073/pnas.2300963120.
Many caveats still apply. LLMs are far from perfect, and I am still struggling with their immediate and eventual impacts on science (see
prior thread). My goal in the current thread is to think about them as cognitive artifacts instead.
For cognitive scientists interested in the emergence of intelligent behavior, LLMs suggest that some wide range of interesting adaptive behaviors can emerge given enough scale. Obviously, there's huge debate over what counts as intelligent, and I'm not going to solve that here.
But: for my money, we start seeing *really* interesting behaviors at the scale of GPT3. Prompting for few shot tasks felt radically unexpected and new, and suggested task abstractions underlying conditional language generation. At what scale do you see this?
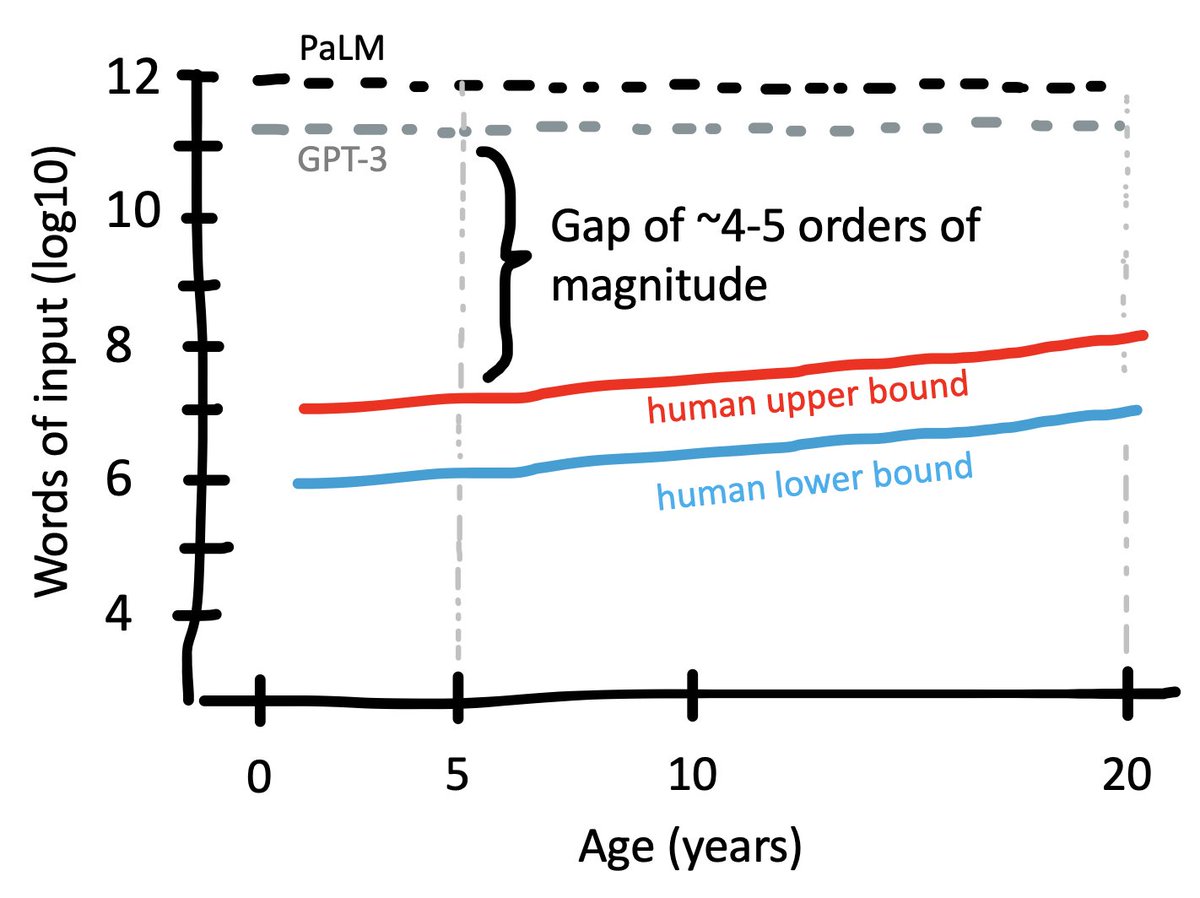
GPT-3 was trained on 500 billion tokens (= .75 words). So that gives us ~4e11 words. PaLM and Chinchilla are both trained on around 1e12 words. We don't know the corpus size for GP4-4 (!?!). How do these numbers compare with humans?
Let’s start with an upper bound. A convenient approximation is 1e6 words per month for an upper bound on spoken language to a kid (
arxiv.org/pdf/1607.08723…, appendix A or
pnas.org/doi/abs/10.107…). That's 2e8 words for a 20 year old. How much could they read?
Assume they start reading when they’re 10, and read a 1e5-word book/week. That’s an extra 5e6 million words per year. Double that to be safe and it still only gets us to 3e8 words over 10 years.
Now let's do a rough lower bound. Maybe 1e5 words per month for kids growing up in a low-SES environment with limited speech to children (
onlinelibrary.wiley.com/doi/epdf/10.11…). We don't get much of a literacy boost. So that gives us 5e6 by age 5 and 2e7 by age 20.
That "lower bound" five year old can still reason about novel tasks based on verbal instructions - especially once they start kindergarten!
The take-home here is that we are off by 4-5 orders of input magnitude in the emergence of adaptive behaviors.
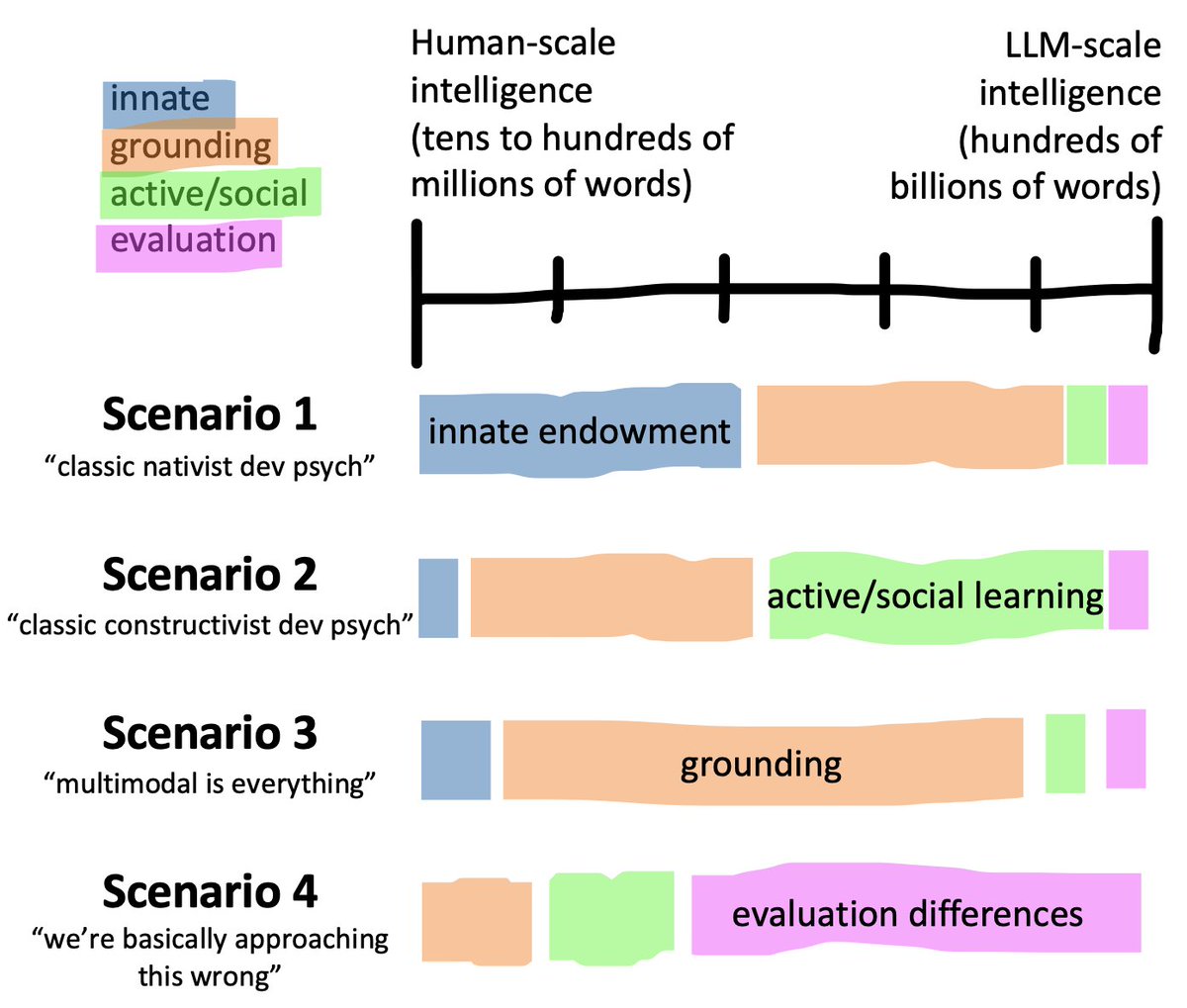
The big cognitive science question is - which factors account for that gap? I'll think about four broad ones.
Factor 1: innate knowledge. Humans have SOME innate perceptual and/or conceptual foundation. The strongest version posits "core knowledge" of objects, agents, events, sets, etc. which serve to bootstrap further learning. People disagree about whether this is true.
Factor 2: multi-modal grounding. Human language input is (often) grounded in one or more perceptual modalities, especially for young children. This grounding connects language to rich information for world models that can be used for broader reasoning.
Factor 3: active, social learning. Humans learn language in interactive social situations, typically curricularized to some degree by the adults around them. After a few years, they use conversation to elicit information relevant to them.
Factor 4: evaluation differences. We're expecting chatGPT to reason about/with all the internet's knowledge, and a five year old just understand a single novel theory of mind or causal reasoning task. Is comparison even possible?
So of course I don't know the answer! But here are a few scenarios for thinking this through. Scenario 1 is classic nativist dev psych: innate endowment plus input make the difference. You use core knowledge to bootstrap concepts from your experience.
Scenario 2 is more like modern rational constructivism. Grounded experience plus a bunch of active and social learning allow kids to learn about the structure of the world even with limited innate knowledge.
I hear more about Scenario 3 in the AI community - once we ground these models in perceptual input, it's going to be easier for them to do common-sense reasoning with less data. And finally, of course, we could just be all wrong about the evaluation (Scenario 4).
As I said, I don't know the answer. But this set of questions is precisely why challenges like BabyLM are so important (
babylm.github.io).